Wet Lab Protocols
Science doesn’t need to be so hard! Most protocols are far from proprietary but finding concise instructions is often difficult. We share most of our protocols so they can hopefully be helpful for other microbiome researchers! See the Lab’s Protocol GitHub Repository
QIIME2R
QIIME2 is one of the most commonly used environments for processing microbiome data; however, it’s use of artifacts limits its direct usage in R. qiime2R facilitates the direct import of QIIME2 artifacts to R sessions for downstream analysis. Find installation and usage instructions here or download from github.
Amplicon Sequencing and Analysis
From feces to feature table. A one-stop shop for preparing sequencing and analyzing libraries with the help of the Opentrons OT-2 liquid handler. See GitHub Repository.
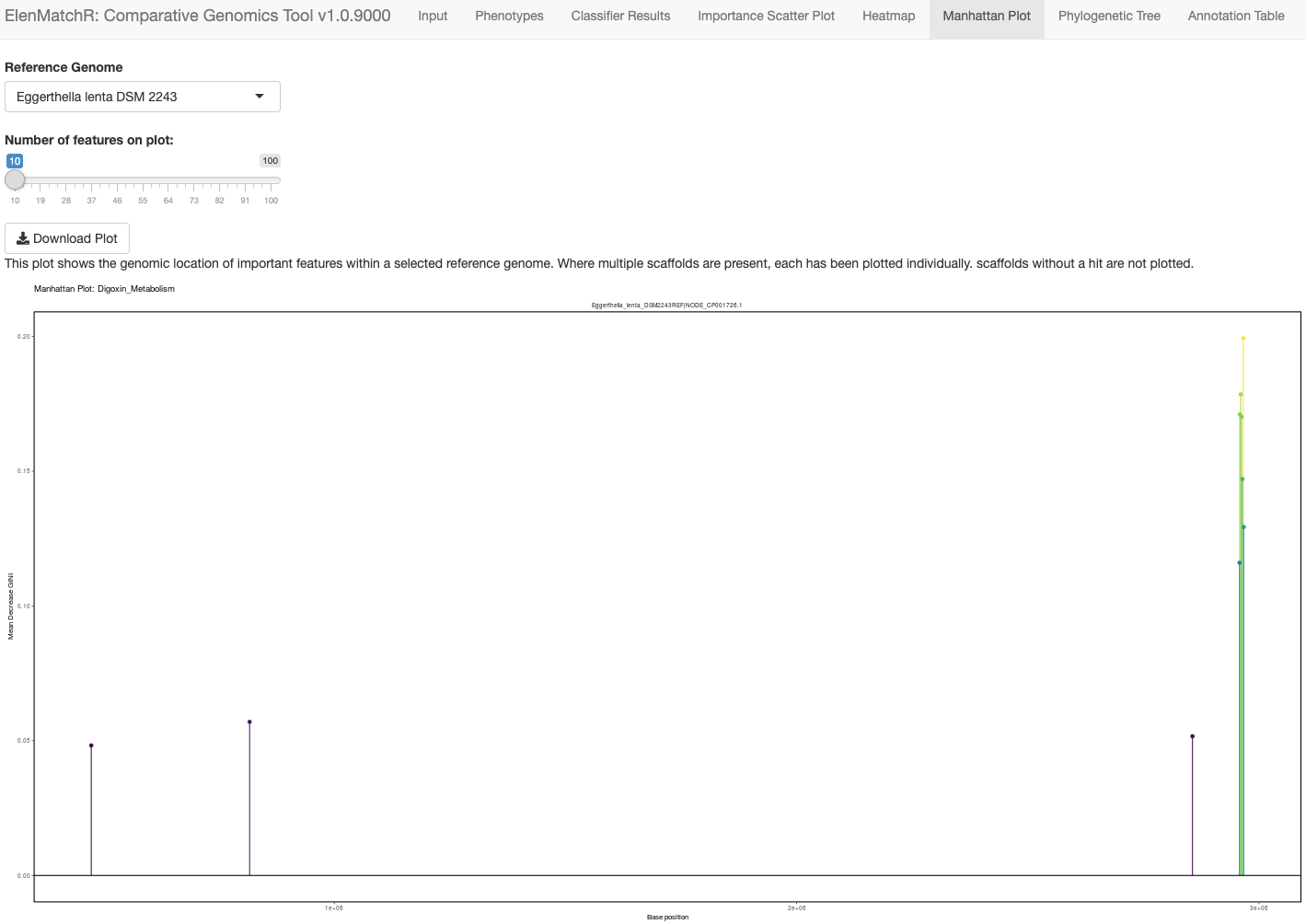
ElenMatchR
A paired strain-library and comparative genomics tool for discovering microbial enzymes doing everything from drug metabolism to immune modulation. Strains are available from the German Culture Collection (DSMZ) and the GUI is available from GitHub.
High Fat Diets and the Microbiome
Diet is one of the major determinants of microbiome structure and fat is bad for your microbiome… right? As a component of a recent meta-analysis of the effect of high-fat diet on the mouse and human gut microbiome (Bisanz et al., Cell Host & Microbe 2020), we established a pipeline for meta-analysis and released the code and datasets.